Eco-evolutionary trajectories of host-microbiome interactions
I integrate concepts from microbial ecology, evolution, genetics, and genomics in my research to uncover eco-evolutionary trajectories of host-microbiome interactions and understand the ecological processes that drive these reciprocal interactions. I integrate a variety of omics tools and evolutionary experiments to:
A) Explore reciprocal coevolution and co-speciation of microbiomes and their host.
B) Identify host/microbial genomic signatures associated with their co-phylogeny success.
C) Explore coevolutionary conflict host-microbiome and pathogens.
In my research, I adopt an environmental approach, acknowledging that microbial ecology and evolution occur across diverse habitats and ecosystems. My investigations encompass distinct ecosystems, including the intricate host-associated microbial communities of humans, plants, and freshwater environments. By embracing a holistic approach, I aim to explore the consistency of eco-evolutionary dynamics of microbiomes across diverse ecological niches.

Project 1. Reciprocal coevolution and co-speciation of microbiome with their host.
Exploring the dynamic interplay between microbiomes and their host: a two-way conversation. Our understanding of adaptation between organisms and their associated microbiota is limited, especially at the genomic level. For example, still is not clear how hosts can have a selective effect on microbial communities, especially at the strain level to select between the beneficial strains (e.g., SCF producer) and the strains that do not have the traits. To tackle this issue, I propose to look at this in three classes of effect:
Microbe-to-host. One of the factors that can change host gene expression is microbiota. Microbiome-induced regulatory changes can have instant effects on host health and physiology, as they change metabolic and immunological pathways. I am planning to modulate the host microbiome, and reciprocally measure host and microbiome gene expression. This will allow me to identify possible microbial and host genes involved in host-microbiota recognition, host-microbiome transcription variation, and possible metabolic exchanges.
Host-to-microbiome. Host genetic control over the microbiome refers to genetic variants responsible for the selection for or against particular microbes by the host. Genetic association between host genetic variants and microbial structural variations may help us understand the mechanisms underlying the symbiotic relationship between host and their microbiome.
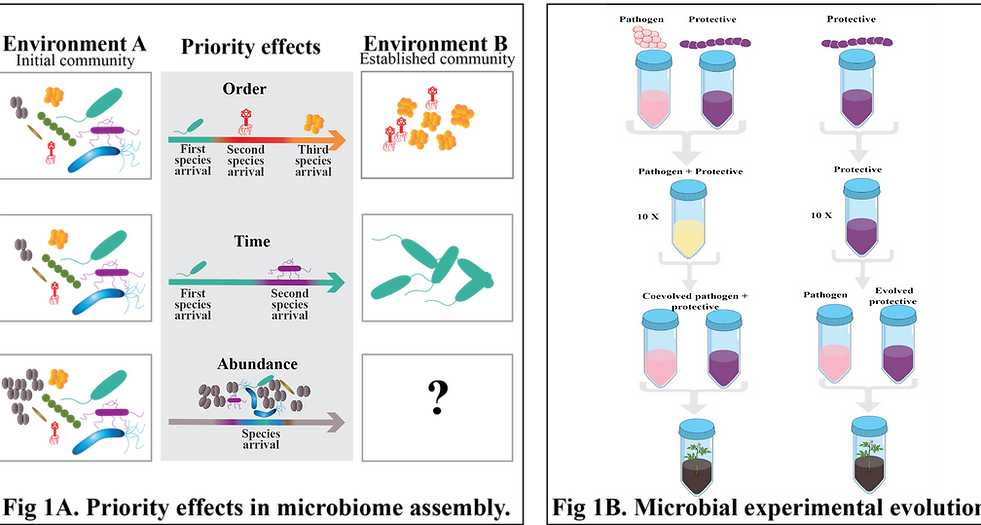
Microbe-microbe. The interactions between microbes strongly influence the presence or absence of other microorganisms in the community and set the overall composition, stability and biodiversity of microbial ecosystems. I am planning to use synthetic microbial communities composed of different numbers of bacterial taxa (thus absolute number) under different environmental conditions and test patterns and processes of microbial community assembly including priority effects, microbial interactions and stability under biotic stress (e.g., pathogens outbreak), and microbial population dynamic using gLV and other microbial models.
Interested to read more? Check out our published work in the Journal of Science of The Total Environment.

Project 2. Coevolutionary conflict of host–pathogens.
The Red Queen hypothesis is centered on the concept of antagonistic coevolution, or host-pathogen coevolution, and presumes that adaptations that increase the fitness of one species will decrease the fitness of another and thus drive selection. Pathogens can evolve mechanisms to evade recognition by the host immune system, such as antigenic variation or the secretion of immunosuppressive molecules. In response, hosts may evolve more sophisticated immune recognition systems or behavioral adaptations to avoid infection.
Microbial coevolution in the presence of pathogens. During host-symbionts-pathogen interactions, continuous coevolution of the host defences, adaptation and evolution of microbiota (symbionts and pathogens), and pathogens evasion take place. Symbionts can control pathogens by competing for nutrients, niche exclusion, and induction of host immune response, collectively known as colonization resistance. Pathogens can also evolve to expand their population by increasing their virulence or less virulent to establish themselves and expand the pathogen population. This means that strains that were once pathogenic may evolve to become neutral or strains that were once neutral may evolve to provide substantial benefits. However, the exact mechanism behind microbial response to pathogens is not clear. I am planning to use multi-omics approaches and experimental evolution to analyze microbial coevolution in the presence of pathogens at genomics, transcriptomics, and metabolomics. Beneficial or deleterious mutations can arise, and using advanced genomics tools such as metagenomics can help us to identify those changes.
Coevolutionary conflict of pathogens with their hosts. Coevolution between hosts and pathogens is universal and often is associated with rapid evolutionary change thus maintaining diversity. Genome rearrangements, altering its antigens, resistance to antibacterial effectors, and mimicking host cells through the acquisition of host cell genes are some of the mechanisms that pathogens can evade from recognition by the host immune system. Hosts also evolve to escape from pathogens. By comparing sequence similarities between host and their pathogens, I aim to find genes that have similarities to their host. Moreover, I am comparing evolved pathogens versus ancestral to find possible genome rearrangements and determine which genes have higher genome rearrangements in pathogens and if those genes are under selection. Ultimately, this knowledge will teach us how host pathways are manipulated and how infections may be tackled.
Pathogens coevolution through co-infection. Co-infections by several microbial pathogens (strains from the same species or different species) are common in the wild. Pathogens that share a common host could interact with the host or between them directly, indirectly, or both. These interactions could have several outcomes for both host and pathogens. For example, stronger symptoms (and higher virulence) under coinfection have frequently been reported. On the other hand, antagonisms in co-infections (milder symptoms) have also been described. Coinfection can also have a drastic impact on the evolution of pathogen populations, from the generation of novel genetic combinations to its potential role in driving evolution through natural selection. Through co-infection experiments, I am trying to find how pathogens are responding to their competitors. This knowledge will help us to understand the pathogen-pathogen interactions and use some of their antagonism responses (e.g., toxins produced by bacteria, or antibiotics produced by fungi in fungi-bacteria co-infections) as a sustainable way of disease prevention.